MARS (Microarray Analysis and Retrieval System)
provides a comprehensive MIAME supportive suite for storing,
retrieving, and analyzing multi color microarray data. The
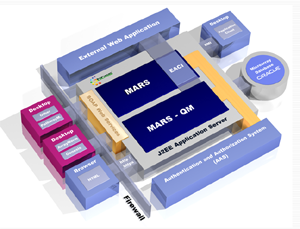
system comprises a lab notebook, a laboratory information
management system (LIMS), a quality control management,
as well as a sophisticated user management system. MARS is
fully integrated into an analytical pipeline of microarray image
analysis, normalization, gene expression clustering, and
mapping of gene expression data onto biological pathways.
The incorporation of ontologies and the use of MAGE-ML enables an export
of studies stored in MARS to public repositories and other databases accepting
these documents. Its unique fusion of using Web based and standalone applications
connected to the latest J2EE application server technology has been tailored
to serve the specific needs of microarray based research projects.
The MARS web application can be found under http://mars.genome.tugraz.at.
For testing you can login to the MARS playground [user: demo password: demo] |
| Maurer M, Molidor R, Sturn A, Hartler J, Hackl H, Stocker G, Prokesch A, Scheideler M, Trajanoski Z. MARS: microarray analysis, retrieval, and storage system. BMC Bioinformatics. 6: 101-101 (2005)
PM:15836795
|














