Cross-Validation Explained
There are two types of cross-validation for the classification accuracy estimation:
- K-fold cross-validation
- Leave-one-out cross-validation
They are discussed in the subsections below.
K-fold Cross-Validation
One iteration of the K-fold cross-validation is performed in the following way:
First, a random permutation of the sample set is generated and partitioned
into K subsets ("folds") of about equal size.
Of the K subsets, a single subset is retained as the validation data for testing
the model (this subset is called the "testset"),
and the remaining K - 1 subsets together are used as training data ("trainset").
Then a model is trained on the trainset and its accuracy is evaluated on the testset.
Model training and evaluation is repeated K times,
with each of the K subsets used exactly once as the testset.
The case of a 5-fold cross-validation with 30 samples is illustrated
in the picture below:
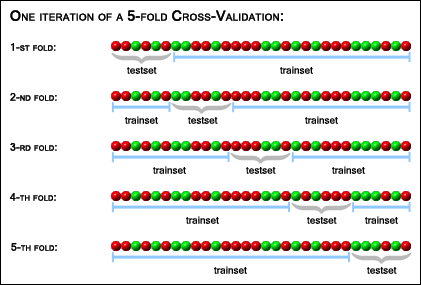
The resulting accuracy estimation depends on the random permutation that was generated
in the beginning of the process, because it affects the way the sample set is partitioned.
Therefore to obtain a more exact estimation of the accuracy it makes sense to repeat
cross-validation several times and take the average of all the accuracy estimations
obtained after each iteration as the resulting accuracy estimation.
Leave-one-out Cross-Validation
As the name suggests, leave-one-out cross-validation involves using a single
sample from the original sample set as the validation data, and the remaining samples
as the training data. This is repeated such that each sample in the sample set is used
exactly once as the validation data.
This is the same as K-fold cross-validation where K is equal to the number of
samples in the sample set.
There is no need in generating random permutations for leave-one-out cross-validation
and in repeating it, because the training and validation datasets for each of the folds
are always the same, and therefore the result of the accuracy estimation is determined.
ProClassify User's Guide
Copyright © 2005-2006
Institute for Genomics and Bioinformatics -
Graz University of Technology
Department of Information Design -
FH JOANNEUM - Graz University of Applied Sciences

